Geographic distribution
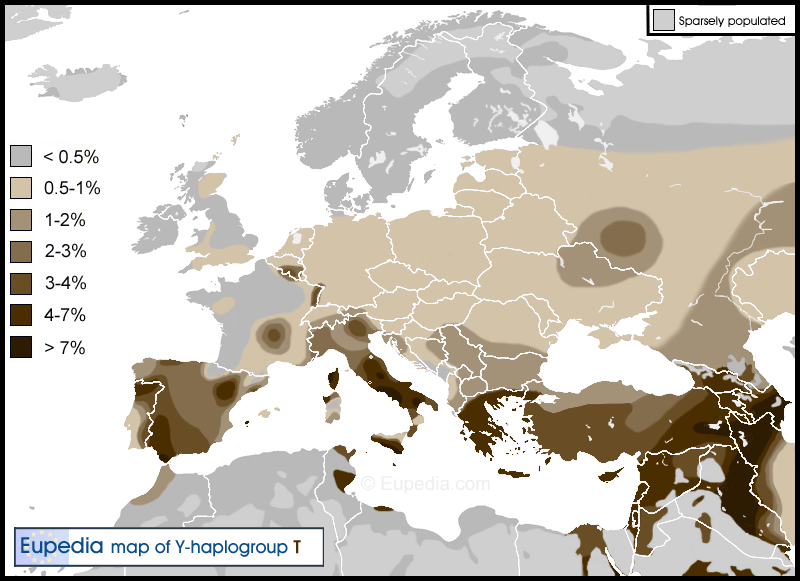
Haplogroup T is one of the most widely dispersed paternal lineages in the world. In Europe, it makes up only 1% of the population on most of the continent, except in Greece, Macedonia and Italy where it exceeds 4%, and in Iberia where it reaches 2.5%, peaking at 10% in Cadiz and over 15% in Ibiza. The maximal worldwide frequency for haplogroup T is observed in East Africa (Eritrea, Ethiopia, Somalia, Kenya, Tanzania) and in the Middle East (especially the South Caucasus, southern Iraq, south-west Iran, Oman and southern Egypt), where it accounts for approximately 5 to 15% of the male lineages. Over 50% of haplogroup T has been reported in some tribes in northern Somalia and Djibouti. Another hotspot are the Fulani people of Cameroon (18%). Besides these regions and Europe, T is found in isolated pockets as far as Zambia, South Africa, India, Central Asia and Northeast Asia, including southern Siberia, Mongolia (2%) and northern China (1%).
Distribution of haplogroup T in Europe, the Middle East and North Africa
Phylogeny of T1a
If you are new to genetic genealogy, please check our Introduction to phylogenetics to understand how to read a phylogenetic tree.


Origins & History
Haplogroup T emerged from haplogroup K, the ancestor of most of the Eurasian haplogroups (L, N, O, P, Q, R and T), some time between 45,000 and 35,000 years ago. The vast majority of modern members of haplogroup T belong to the T1a branch, which developed during the late glacial period, between 25,000 and 15,000 years ago, possibily in the vicinity of the Iranian Plateau.
Although haplogroup T is more common today in East Africa than anywhere else, it almost certainly spread from the Fertile Crescent with the rise of agriculture. Indeed, the oldest subclades and the greatest diversity of T is found in the Middle East, especially around the Fertile Crescent. Lazaridis et al. (2016) identified one carrier of haplogroup T among the remains of the Pre-Pottery Neolithic B site in Jordan. A T1a sample was also found in the Early Neolithic Linear Pottery (LBK) culture in Germany by Mathieson et al. (2015). By the end of the last glacial period, 12,000 years ago, haplogroup T had already differentiated into subclades such as T1a1a, T1a2, T1a3a and T1a3b. Deeper subclades developed in the Near East during the Early Neolithic period for several millennia before early farmers started expanding beyond the Near East.
Neolithic colonisation of the Arabian peninsula and East Africa
The higher frequency of T in East Africa would be due to a founder effect among Neolithic farmers or pastoralists from the Middle East. One theory is that haplogroup T spread alongside J1 as herder-hunters in the Pre-Pottery Neolithic period, leaving the Zagros mountains between 9,000 and 10,000 BCE, reaching the Egypt and the southern Arabian peninsula around 7,000 BCE, then propagating from there to the Horn of Africa, and later on to Madagascar. However, considering that J1 peaks in Yemen and Sudan, while T1 is most common in southern Egypt, Eritrea and Somalia, the two may not necessarily have spread together. They might instead have spread as separate nomadic tribes of herders who colonised the Red Sea region during the Neolithic, a period than spanned over several millennia. Nevertheless both are found in all the Arabian peninsula, all the way from Egypt to Somalia, and in Madagascar. This contrasts with other Near Eastern haplogroups like G2a and J2, which are conspicuously absent from East Africa, and rare in the Arabian peninsula. Nowadays, T1a subclades dating from the Neolithic found in East Africa include Y16247 (downstream of CTS2214) and Y16897. Other subclades dating from the Bronze Age (see below) are present as well, such as Y15711 and Y21004, both downstream of CTS2214.
Neolithic colonisation of Southern Europe
Ancient DNA tests have revealed that the majority of Early Neolithic farmers who colonised Europe belonged to Y-haplogroup G2a, with a minority of other haplogroups such as C1a2, E1b1b, H2, I2, J2 and T1a. In fact, at present haplogroup T has only been identified in remains from the Linear Pottery (LBK) culture in Germany. Nevertheless, less than 100 Neolithic European Y-DNA samples have been tested to date, so it is too early to tell if G2a was dominant in all Neolithic European cultures or only in south-east and central Europe.
Nowadays haplogroup T1a is far more common in Mediterranean Europe than elsewhere on the continent. This suggests that the bulk of T1a farmers may have followed a different route from the Near East, either by boat along the coasts, and/or via North Africa. In the first hypothesis, T1a farmers would have been associated with the Cardium Pottery culture (5000-1500 BCE). However, none of the samples from this culture or from later Neolithic cultures in Iberia or Italy have yielded any T1a samples (once again G2a is the dominant lineage). The North African route is therefore more likely and would explain why the T1a subclades found in Europe, notably T1a1a1a1 (CTS2214), T1a1a1b (Y6671), T1a1a2 (Y16897), T1a2a1 (Y6055) and T1a2b1 (P322) are all found especially in the Southwest Asia (South Levant and Arabian penisula) and Europe. Little data is available for North Africa, but some of these subclades have been confirmed in Egypt (Y6671>Y12643), Libya (Y6671>Y22559) and Morocco (Y6671>Y12643).
Furthermore, within Europe the frequency of T1a mirrors that haplogroup J1, just as they do in the Arabian peninsula. In Europe, both haplogroups are most common in the mountainous parts of the southern Balkans, the central and southern Apennines in Italy, Sicily, the Massif Central in France (Auvergne), and south-western Iberia. All these regions would have been better suited for goat and sheep herding than for cereal cultivation in the Neolithic period. Mountainous regions allow herders to practice transhumance, i.e. the seasonal movement of people with their livestock between fixed summer and winter pastures.
Neolithic colonisation of Northeast Europe and Siberia
T1a2 (L131) has been found as far east as the Volga-Ural region of Russia and Xinjiang in north-west China. This branch probably penetrated into the Pontic-Caspian Steppe during the Neolithic (perhaps alongside J2b) and became integrated to the indigenous R1a peoples before their expansion to Central Asia during the Bronze Age (=> see R1a-Z93). Nour Mousa's doctoral thesis reported the presence of haplogroup K (xN1c1-TAT, O-M175, P-M45) among Early Neolithic inhabitants of the Lake Baikal region in southern Siberia. That sample was not tested for haplogroup T, but it very probably is T1a considering its presence in the region today and the lack of other plausible alternative (besides K* and N*).
Haplogroup T has been found at a relatively high frequency among the Tatars (5%) and Maris (2%) of the Volga-Ural region as well as in north-west Russia (3%) suggesting that it may have been one of the principal lineages bringing the Neolithic to Uralic-speaking population. Autosomal DNA tests have also identified unusually high percentages of Southwest Asian admixtures among the Finns (1 to 2.5%) and Lithuanians (1.5%), who otherwise lack West Asian or Caucasian admixture and possess hardly any Middle Eastern Y-DNA. This Southwest Asian admixture could be the trace of T lineages absorbed during the Neolithic.
Kura-Araxian expansion during the Bronze Age
The P77 and CTS6507 branch underwent a major expansion during the Early Bronze Age, from approximately 2500 BCE. The phylogeny suggests that this expansion took place from the South Caucasus region, including the Armenian Highlands, and spread in various directions around the Middle East and Europe. The European branch appears to have propagated through a Mediterranean route to Greece, Italy (including Sicily and Sardinia) and Iberia. Historically the Kura-Araxes culture is the best match for this expansion. While the Proto-Indo-Europeans (haplogroups R1a and R1b) were expanding from the Pontic-Caspian Steppe to central and northern Europe and Central Asia, the Kura-Araxes people, on the other side of the Caucasus, also developed a contemporary Bronze Age culture that expanded across West Asia, and possibly as far east as Pakistan and India. The Minoans, Europe's oldest proper civilisation (as opposed to archeological culture), could be an offshoot from that Kura-Araxes expansion. Kura-Araxian men would have belonged primarily to Y-haplogroup J2a1, but also to a lower extent to G2a-L293, G2a-M406, J1-Z1828, L1b, T1a-P77, and R1b1-L278.
Ancient civilisations associated with Y-haplogroup T
During the Copper and Bronze Ages haplogroup T would have been an important (though probably not dominant) lineage among ancient peoples such as Sumerians, the Babylonians and the Assyrians.
The higher than average frequencies of haplogroup T in places like Cyprus, Sicily, Tunisia, Ibiza, Andalusia and the northern tip of Morocco suggest that haplogroup T could also have been dispersed around the Mediterranean by the Phoenicians (1200-800 BCE), and that ancient Phoenicia seemingly had a higher incidence of T than Lebanon does today (5%).
The maternal lineages (mtDNA) corresponding to haplogroup T
One way of determining the original maternal lineages that could have accompanied the Early Neolithic farmers belonging to Y-haplogroup T is to compare the mtDNA haplogroups found in regions with high frequencies of haplogroup T today. It is particularly useful to look at very distant regions and ethnic groups that do not share other paternal lineages than T. Fortunately Neolithic T tribes have dispersed far and wide in all directions from the Near East, colonising Europe, East Africa, parts of India and even Siberia. Analysing the mtDNA haplogroups of East African countries with high frequencies of T, such as Ethiopia, Somalia or parts of Madagascar, and comparing them with ancient samples of Neolithic farmers, the best matches are mt-haplogroups HV, N1a and U3.
Famous individuals

The testing of Thomas Jefferson's DNA revealed that the third US president belonged to haplogroup T. Descendants of his father, Colonel Peter Jefferson, tested on the Jefferson DNA Project and belonged to haplogroup T1a1a-L208 > CTS11451 > Y6671.
Follow-up
Ask your questions and discuss about haplogroups on the Forum