Mal'ta boy had autosomal genes present in populations with Y-haplogroups M, P, Q & R
Two weeks ago, Raghavan et al. published a paper on the genome of an Upper Palaeolithic Siberian individual, known as the Mal'ta boy. It is by far the oldest human genome tested to date.
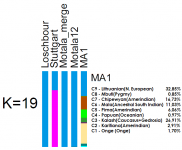
The authors reported that the autosomal admixture of that 24,000-year-old individual was a blend of European, South Asian, Amerindian with a little bit of Papuan. I don't know if anybody mentioned this before, but it strikes me that this particular admixture could in fact be the original source DNA representing descendants of haplogroup MP. What links Europeans and South Asian is essentially haplogroup R. Amerindians are Q and Papuans belong too M.
It has recently been found that haplogroup M fits in between NO and P in the Y-chromosomal phylogeny. In other words, there was once a haplogroup MP, which split into M and P, then P split into Q and R.
Since the Mal'ta boy belongs to R*, it makes sense that his autosomal genes should be closest to the populations with the highest percentages of haplogroup R today, namely (North) Europeans and South Asians. And indeed his genome resembles at 71% that of Europeans and South Asians. The second closest group is Q, which is mostly Amerindians. 26% of his genome matches that of Amerindian. Surely this is something inherited from the common roots of Q and R, which had split from each others only a few millennia before the Mal'ta boy's lifetime.
The Mal'ta boy also shared 4% of similarity with modern Papuans, which may come as a surprise at first, until one realises the close phylogenetic relationship between the dominant Papuan paternal lineage (M) with Q and R. However since the split happened longer ago, the genetic similarity is more limited than with Q. Besides, there is a good chance that modern Papuans only have a small percentage of Eurasian genes themselves, and that the original carriers of Y-haplogroup M intermingled with a lot of other populations on their long journey from Central/North Asia to Papua. It is possible that only a small group of men belonging to haplogroup M came to replace the older native paternal lineages of New Guinea (C-RPS4Y711 and C-P55), just as O replaced them in East Asia and R in South Asia and Europe. Even Q might represent the post-Clovis migration that replaced older lineages (C3 ?) among Amerindians.
So we shouldn't see the Mal'ta boy as a multi-hybrid of European, South Asian and Siberian/Amerindian ethnicities, but rather as an example of the source population which invaded those geographic regions and hybridised with the natives there. It would be interesting to use the Mal'ta boy's genome as a reference population and see how much of modern populations inherited from the original PQR people, instead of looking at it the other way round. There might be a correlation between the percentage of similarity with his autosomal genes and the frequency of haplogroups Q and R in modern populations. Nonetheless I would expect that autosomal DNA got progressively diluted along the way as R people moved into Europe and South Asia, so the maximum percentage of similarity would probably lie between Bactria and Northwest India and in Eastern Europe.
Two weeks ago, Raghavan et al. published a paper on the genome of an Upper Palaeolithic Siberian individual, known as the Mal'ta boy. It is by far the oldest human genome tested to date.
The authors reported that the autosomal admixture of that 24,000-year-old individual was a blend of European, South Asian, Amerindian with a little bit of Papuan. I don't know if anybody mentioned this before, but it strikes me that this particular admixture could in fact be the original source DNA representing descendants of haplogroup MP. What links Europeans and South Asian is essentially haplogroup R. Amerindians are Q and Papuans belong too M.
It has recently been found that haplogroup M fits in between NO and P in the Y-chromosomal phylogeny. In other words, there was once a haplogroup MP, which split into M and P, then P split into Q and R.
Since the Mal'ta boy belongs to R*, it makes sense that his autosomal genes should be closest to the populations with the highest percentages of haplogroup R today, namely (North) Europeans and South Asians. And indeed his genome resembles at 71% that of Europeans and South Asians. The second closest group is Q, which is mostly Amerindians. 26% of his genome matches that of Amerindian. Surely this is something inherited from the common roots of Q and R, which had split from each others only a few millennia before the Mal'ta boy's lifetime.
The Mal'ta boy also shared 4% of similarity with modern Papuans, which may come as a surprise at first, until one realises the close phylogenetic relationship between the dominant Papuan paternal lineage (M) with Q and R. However since the split happened longer ago, the genetic similarity is more limited than with Q. Besides, there is a good chance that modern Papuans only have a small percentage of Eurasian genes themselves, and that the original carriers of Y-haplogroup M intermingled with a lot of other populations on their long journey from Central/North Asia to Papua. It is possible that only a small group of men belonging to haplogroup M came to replace the older native paternal lineages of New Guinea (C-RPS4Y711 and C-P55), just as O replaced them in East Asia and R in South Asia and Europe. Even Q might represent the post-Clovis migration that replaced older lineages (C3 ?) among Amerindians.
So we shouldn't see the Mal'ta boy as a multi-hybrid of European, South Asian and Siberian/Amerindian ethnicities, but rather as an example of the source population which invaded those geographic regions and hybridised with the natives there. It would be interesting to use the Mal'ta boy's genome as a reference population and see how much of modern populations inherited from the original PQR people, instead of looking at it the other way round. There might be a correlation between the percentage of similarity with his autosomal genes and the frequency of haplogroups Q and R in modern populations. Nonetheless I would expect that autosomal DNA got progressively diluted along the way as R people moved into Europe and South Asia, so the maximum percentage of similarity would probably lie between Bactria and Northwest India and in Eastern Europe.
Last edited: