Install the app
How to install the app on iOS
Follow along with the video below to see how to install our site as a web app on your home screen.
Note: This feature currently requires accessing the site using the built-in Safari browser.
You are using an out of date browser. It may not display this or other websites correctly.
You should upgrade or use an alternative browser.
You should upgrade or use an alternative browser.
GEDmatch, list of ancient samples with kit numbers.
torzio
Regular Member
- Messages
- 3,991
- Reaction score
- 1,245
- Points
- 113
- Location
- Eastern Australia
- Ethnic group
- North East Italian
- Y-DNA haplogroup
- T1a2 - SK1480
- mtDNA haplogroup
- H95a
Lukas
Regular Member
- Messages
- 328
- Reaction score
- 240
- Points
- 43
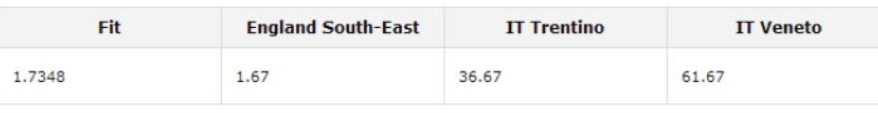
Yes, sorry. Trentino.
torzio
Regular Member
- Messages
- 3,991
- Reaction score
- 1,245
- Points
- 113
- Location
- Eastern Australia
- Ethnic group
- North East Italian
- Y-DNA haplogroup
- T1a2 - SK1480
- mtDNA haplogroup
- H95a
Yes, sorry. Trentino.
So, you can check more for trentino ?
I have recently joined one of their sites and i have 2 x dna matches in val di non and 2 x matches in Fonzaso area
Tamakore
Regular Member
- Messages
- 181
- Reaction score
- 62
- Points
- 28
- Location
- Wellington
- Ethnic group
- Maori Irish French Scottish English
- Y-DNA haplogroup
- R1b-L21 DF5>BY154246
- mtDNA haplogroup
- J1c3b2
When will the Dzudzuana samples be uploaded to GEDmatch, that's what I'd like to know? A calculator based on Dzudzuana would also be good.
Last edited:
Carlos
Banned
- Messages
- 2,647
- Reaction score
- 700
- Points
- 0
- Y-DNA haplogroup
- E-V22/YF66572
- mtDNA haplogroup
- J1c5c1
Comparing KitXXXXXX [FTDNA] and M276583 (la Brana WHG) [Migration - V3 - M]
Precision: 30.0
cM threshold: 2.0
Maximum cM: No Limit
Gap threshold: 2.0 cm's
All SNPs used.
Total cM: 10.02
Largest segment cM: 2.91
Total segments: 4
Total gap-induced breaks: 2
Max gap: 2.16
..... gap: 44881336-70984372 on chromosome 9
Top 10 Q scores:
Q-score: 25.00
Q-score: 12.00
Q-score: 8.00
Q-score: 0.00
Comparison took 0.888 seconds.
CPU time used: 0.874 cpu seconds.
By putting 2 I have obtained results, in other samples putting 5 I have also obtained.
Minimum segment cM size
to be included in total:
(Leave blank for default value = 7)
-----------------------------------------
I have done this one at random, it has made me funny to see that country so small and flirtatious, nor had I seen it here before.
[SIZE=-1]Software Version Jun 26 2019 00:29:33[/SIZE]
Comparing xxxxxxxxx [FTDNA] and F999918 (Loschbour, Lux., 8ky) [GEDmatch Xfer]
Precision: 30.0
cM threshold: 4.0
Maximum cM: No Limit
Gap threshold: 2.0 cm's
All SNPs used.
Total cM: 12.99
Largest segment cM: 4.56
Total segments: 3
Total gap-induced breaks: 0
---------------------------------
Comparing xxxxxx [FTDNA] and M124870 (I1661 IranChL) [Migration - V3 - M]
Total cM: 4.82
Largest segment cM: 2.47
Total segments: 2
Total gap-induced breaks: 5
Max gap: 2.20
..... gap: 44866028-71040170 on chromosome 9
Top 10 Q scores:
Q-score: 10.00
Q-score: 2.00
-------------------------------------------------
Comparing xxxxxxx [FTDNA] and M690970 (RISE386_Sintashta) [Migration - V4 - M]
Total cM: 3.08
Largest segment cM: 3.08
Total segments: 1
Total gap-induced breaks: 4
Max gap: 2.29
..... gap: 28374012-29211558 on chromosome 15
-----------------------
Comparing Kit xxxxxxxx [FTDNA] and M547763 (I1300 Spain Chalcolithic) [Migration - V3 - M]
Total cM: 12.98
Largest segment cM: 3.48
Total segments: 5
Total gap-induced breaks: 3
Max gap: 2.16
..... gap: 44866028-70984372 on chromosome 9
-------------------------------------------
This sample had already been obtained at MTA
[FONT="]Comparing xxxxxxxx[FTDNA] and M427312 (Ballynahatty IrelandNE) [Migration - V3 - M]
[/FONT][FONT="]Precision: 30.0[/FONT]
[FONT="]cM threshold: 3.0[/FONT]
[FONT="]Maximum cM: No Limit[/FONT]
[FONT="]Gap threshold: 2.0 cm's
[/FONT][FONT="]Total cM: 10.57[/FONT]
[FONT="]Largest segment cM: 3.71[/FONT]
[FONT="]Total segments: 3[/FONT]
[FONT="]Total gap-induced breaks: 0[/FONT]
Precision: 30.0
cM threshold: 2.0
Maximum cM: No Limit
Gap threshold: 2.0 cm's
All SNPs used.
| Chr | B37 Start Pos'n | B37 End Pos'n | SNPs | Centimorgans (cM) | Q |
| 2 | 8,750,673 | 9,831,821 | 170 | 2.78 | 0.0 |
| Chr | B37 Start Pos'n | B37 End Pos'n | SNPs | Centimorgans (cM) | Q |
| 12 | 6,640,267 | 7,631,883 | 199 | 2.32 | 12.0 |
| Chr | B37 Start Pos'n | B37 End Pos'n | SNPs | Centimorgans (cM) | Q |
| 14 | 30,055,573 | 31,265,509 | 178 | 2.01 | 25.0 |
| Chr | B37 Start Pos'n | B37 End Pos'n | SNPs | Centimorgans (cM) | Q |
| 18 | 7,096,977 | 7,688,826 | 172 | 2.91 | 8.0 |
Total cM: 10.02
Largest segment cM: 2.91
Total segments: 4
Total gap-induced breaks: 2
Max gap: 2.16
..... gap: 44881336-70984372 on chromosome 9
Top 10 Q scores:
Q-score: 25.00
Q-score: 12.00
Q-score: 8.00
Q-score: 0.00
Comparison took 0.888 seconds.
CPU time used: 0.874 cpu seconds.
By putting 2 I have obtained results, in other samples putting 5 I have also obtained.
Minimum segment cM size
to be included in total:
(Leave blank for default value = 7)
-----------------------------------------
I have done this one at random, it has made me funny to see that country so small and flirtatious, nor had I seen it here before.
[SIZE=-1]Software Version Jun 26 2019 00:29:33[/SIZE]
Comparing xxxxxxxxx [FTDNA] and F999918 (Loschbour, Lux., 8ky) [GEDmatch Xfer]
Precision: 30.0
cM threshold: 4.0
Maximum cM: No Limit
Gap threshold: 2.0 cm's
All SNPs used.
Total cM: 12.99
Largest segment cM: 4.56
Total segments: 3
Total gap-induced breaks: 0
---------------------------------
Comparing xxxxxx [FTDNA] and M124870 (I1661 IranChL) [Migration - V3 - M]
Total cM: 4.82
Largest segment cM: 2.47
Total segments: 2
Total gap-induced breaks: 5
Max gap: 2.20
..... gap: 44866028-71040170 on chromosome 9
Top 10 Q scores:
Q-score: 10.00
Q-score: 2.00
-------------------------------------------------
Comparing xxxxxxx [FTDNA] and M690970 (RISE386_Sintashta) [Migration - V4 - M]
Total cM: 3.08
Largest segment cM: 3.08
Total segments: 1
Total gap-induced breaks: 4
Max gap: 2.29
..... gap: 28374012-29211558 on chromosome 15
-----------------------
Comparing Kit xxxxxxxx [FTDNA] and M547763 (I1300 Spain Chalcolithic) [Migration - V3 - M]
Total cM: 12.98
Largest segment cM: 3.48
Total segments: 5
Total gap-induced breaks: 3
Max gap: 2.16
..... gap: 44866028-70984372 on chromosome 9
-------------------------------------------
This sample had already been obtained at MTA
[FONT="]Comparing xxxxxxxx[FTDNA] and M427312 (Ballynahatty IrelandNE) [Migration - V3 - M]
[/FONT][FONT="]Precision: 30.0[/FONT]
[FONT="]cM threshold: 3.0[/FONT]
[FONT="]Maximum cM: No Limit[/FONT]
[FONT="]Gap threshold: 2.0 cm's
[/FONT][FONT="]Total cM: 10.57[/FONT]
[FONT="]Largest segment cM: 3.71[/FONT]
[FONT="]Total segments: 3[/FONT]
[FONT="]Total gap-induced breaks: 0[/FONT]
Anhanguera
Junior Member
- Messages
- 10
- Reaction score
- 17
- Points
- 3
What about the Gedmatch kits of the Roman samples (study "Ancient Rome: A genetic crossroads of Europe and the Mediterranean")? Are they available?
Salento
Regular Member
- Messages
- 5,635
- Reaction score
- 2,838
- Points
- 113
- Ethnic group
- Italian
- Y-DNA haplogroup
- T1a2 - SK1480
- mtDNA haplogroup
- H12a
What about the Gedmatch kits of the Roman samples (study "Ancient Rome: A genetic crossroads of Europe and the Mediterranean")? Are they available?
I know of R1 Kit AG4512653
... but, ... here is a calculator with all of them,
just add your dodecad K7 or K12 coordinates in “TARGET” :
http://vahaduo.genetics.ovh/dodecad-k7b-ancient-vahaduo.htm
http://vahaduo.genetics.ovh/dodecad-k12b-ANCIENT-vahaduo.htm
Anhanguera
Junior Member
- Messages
- 10
- Reaction score
- 17
- Points
- 3
^ Thanks, I knew the results of R1. Thanks for the Vahaduo links too. My interest is also in seeing the Roman samples results on Gedmatch, k15, k13, etc. So many kits, of so many different samples, have been downloaded, and yet nearly all Roman samples are apparently missing. 
Salento
Regular Member
- Messages
- 5,635
- Reaction score
- 2,838
- Points
- 113
- Ethnic group
- Italian
- Y-DNA haplogroup
- T1a2 - SK1480
- mtDNA haplogroup
- H12a
^ Thanks, I knew the results of R1. Thanks for the Vahaduo links too. My interest is also in seeing the Roman samples results on Gedmatch, k15, k13, etc. So many kits, of so many different samples, have been downloaded, and yet nearly all Roman samples are apparently missing.
GedMatch changed its rules, and the raw-data of many ancient samples doesn’t comply with its new requirements.
K13 Ancient (with Romans)
http://vahaduo.genetics.ovh/Eurogenes_k13_ancient-vahaduo.htm
K15 Ancient (with Iron Age Romans)
http://vahaduo.genetics.ovh/k15ancient-vahaduo.htm
Anhanguera
Junior Member
- Messages
- 10
- Reaction score
- 17
- Points
- 3
Thanks for the information. A pity then. Some ancient samples are still running, they are likely from before the rules were changed.
This thread has been viewed 117258 times.